DNA sequencing information has traditionally been elucidated through Sanger sequencing, but Sanger sequencing is slow and limited in the number of sequences that can be processed at one time. In contrast, next generation sequencing (NGS) or high throughput sequencing (HTS) platforms can process millions to billions of DNA fragments simultaneously, whose speed and high-throughput capabilities are exceptional.
NGS sequencing is a sequence-by-synthesis (SBS) technology based on reversible termination sequencing and paired-end sequencing. By using four fluorescently labeled nucleotides to sequence tens of millions of small "clusters" on the surface of the flow cell in parallel, the sequence of tens of millions of DNA molecules can be achieved at one time and efficient and rapid nucleotide data can be generated.

How Does NGS Work?
Currently, Alfa Chemistry has established a strong presence in the sequencing industry, utilizing a platform that focuses on the attachment of single-stranded DNA fragments to a flow cell, followed by bridge amplification of the single-molecule DNA templates and sequencing by synthesis using reversible terminators. The NGS workflow employed by Alfa Chemistry encompasses several key steps.
- Library preparation
- Bridge amplification
- Sequencing
- Data analysis
Library preparation
DNA library construction is the basis of second-generation sequencing technology. Its essence is to convert large DNA fragments of the sample to be measured into small fragments of DNA, and then add adapters to both ends of the small fragment DNA molecules to form effective library molecules. The specific steps are as follows (Fig. 1).
- DNA fragmentation: DNA is first fragmented into smaller input-sized fragments by enzymes or by sonication.
- End repair: The ends of these fragments are repaired.
- Add 3' A tail: Use the Klenow enzyme to add an A base to the 3'end, making it easier to link subsequent primers and adapters.
- Ligate adapters: Specific adapters are ligated to the ends of the fragments, allowing hybridization to the flow cell to occur, including sequencing primer binding sites, tag sequence "index" (barcodes, labeling the adapters in the library) and sequences (P5/P7) complementary to the oligonucleotide sequences fixed in the flow cell.
- Screen fragments: Remove large fragments and various impurities in the system to obtain library fragments with ligated adapters.
- PCR amplification: Amplify the DNA fragment with an adapter using primers complementary to the adapter.
- Quality inspection: Verify whether fragment sizes meet the sequencing requirements by performing a quality inspection.
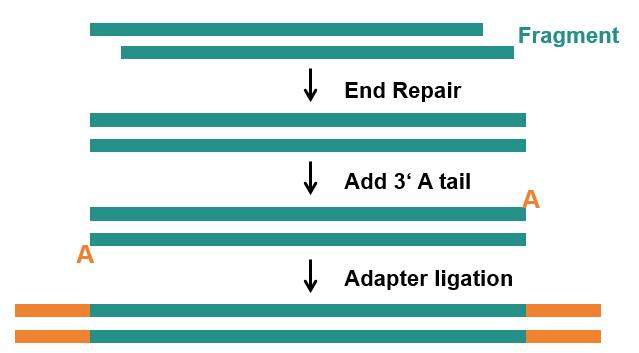 Fig. 1 Diagram of the library construction, including end repair, add 3 ' A tail, and adapter ligation steps.
Fig. 1 Diagram of the library construction, including end repair, add 3 ' A tail, and adapter ligation steps.
Bridge amplification
A flow cell is a hollow glass slide with channels ("lanes"), and is a core element of modern DNA sequencing instruments, aiming at generating clusters of DNA strands for further sequencing and analysis. The flow cell is coated with two types of oligonucleotides (P5' and P7') complementary to the two adapters on the fragment strand, respectively, so that single-stranded, adapter-ligated DNA fragments can attach through hybridization.
The purpose of bridge amplification is to amplify the amount of target DNA so that it eventually reaches the signal intensity required for sequencing reactions. And the bridge amplification PCR process is listed below.
- The sequence on the flowcell surface is used as a template to extend the complementary strand, and then the template strand is washed away, leaving the newly extended reverse strand.
- The reverse strand then folds over and its adapter region hybridizes to the second type of oligo on the flow cell to form single-stranded DNA (ssDNA), and then forms a double-stranded bridge (dsDNA) with the participation of polymerase.
- The double-stranded bridge is denatured and forms two single-stranded copies of DNA, forward and reverse strands, anchored to the flow cell. This denaturation-extension cycle is repeated to form localized DNA clusters on the flow cell.
- After bridge PCR amplification, the reverse strand is washed away, leaving only the forward strand, and block ligation at the 3' end of an oligonucleotide to avoid unnecessary primer binding and amplification.
Sequencing
The prepared library is placed on a sequencer which reads the sequence of each fragment. Modern NGS platforms can process millions to billions of fragments simultaneously, resulting in large amounts of sequencing data.
Data analysis
After the sequencing is completed, the recorded fluorescence signal needs to be processed by specific computer software to obtain the sequence information of DNA.
- Base Calling: The raw signal data from the sequencer is converted into nucleotide sequences (A, T, C, G).
- Read Alignment: The sequences (reads) are aligned to a reference genome or assembled de novo if no reference genome is available.
- Variant Calling: Identifying differences (variants) between the sequenced reads and the reference genome. Variants can include single nucleotide polymorphisms (SNPs), insertions, deletions, and structural variations.
- Quantification (for RNA-Seq): If RNA sequencing is performed, quantification of gene expression levels is done by counting the number of reads that align to each gene.
The NGS workflow is a complex and highly automated process that enables high-throughput and high-resolution analysis of genetic material, revolutionizing fields such as genomics, transcriptomics, and personalized medicine. Unlock the full potential of your research with our comprehensive and cutting-edge NGS services. Our commitment to quality, innovation, and customer satisfaction ensures that you receive the best possible results, every time. Contact us today to discuss your project and discover how we can help you achieve your scientific goals.
