Based on the number and/or assembly of pharmacophores, DNA-encoded libraries (DELs) can be divided into three formats, including single-pharmacophore DELs, dual-pharmacophore DELs, and more recently, trio-pharmacophore DELs (also named triplex DELs), as shown in Fig. 1. Among them, the single-pharmacophore DELs and the dual-pharmacophore DELs are the most common formats of DELs, which are distinguished and classified according to whether the chemical compound is attached to one or two chains of double-stranded oligonucleotides. Besides, trio-pharmacophore DELs, also named triplex DELs, has triplexes of oligonucleotides and is a larger self-assembled DELs than dual-pharmacophore DELs [1].
Alfa Chemistry has established the DELs technology platform, enabling us to provide construction services of DELs in different formats. We are investigating and exploring trio-pharmacophore DELs construction strategies in addition to single- and dual-pharmacophore DELs. If you need construction services of DELs, please feel free to contact us. With our ongoing exploration of science's frontiers, our research team feels confident in providing you with the services you need.
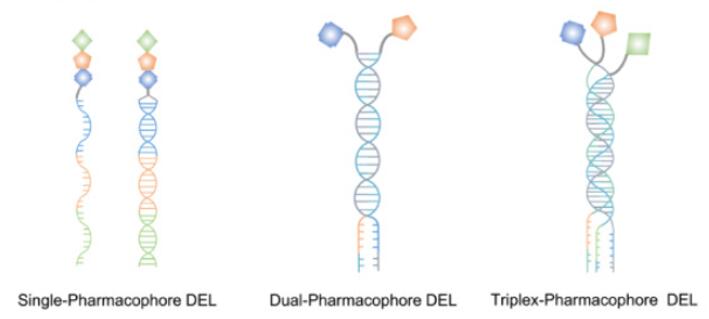 Fig. 1 Schematic representation of three formats of DELs [1].
Fig. 1 Schematic representation of three formats of DELs [1].
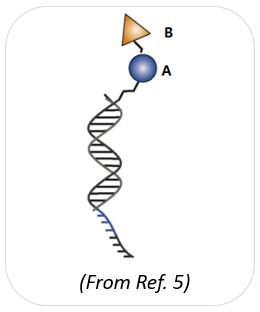
A single-pharmacophore chemical library is a DELs in which individual organic molecules (no matter how complex) are attached to distinctive DNA fragments. In other words, a member of a single-pharmacophore library results from the coupling of a single DNA fragment to a chemical building block. In the case of a two-building block library, the synthesis of single-pharmacophore DELs mainly uses the "split-and-pool" method to generate A x B combinatorial libraries [2].
Alfa Chemistry provides construction services of single-pharmacophore DELs. Please click the link above to view our construction strategies and service process in detail.
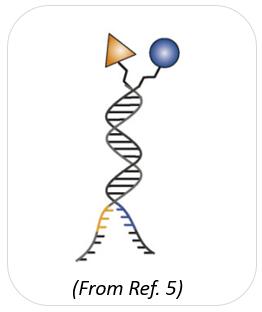
Dual-pharmacophore chemical libraries can be produced by coupling pairs of organic molecules at the extremity of complementary DNA strands. If two sublibraries are constructed, featuring two sets of partially complementary oligonucleotides (each modified with organic molecules), these two sublibraries may then be reassembled (i.e., hybridized) to create a large combinatorial diversity. Notably, each library member can be purified and characterized, making it possible to construct large libraries of high purity. This technology is sometimes referred to as encoded self-assembling chemical (ESAC) library technology. In ESAC libraries, pairs of chemical building blocks are attached to the ends of complementary DNA strands [2,3].
Alfa Chemistry has extensive experience in constructing dual-pharmacophore DELs. Please click the link above to view our construction strategies and service process in detail.
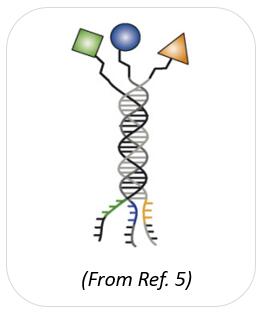
Trio-pharmacophore DELs refers to a triplex library of oligonucleotide conjugates and is an extension of dual-pharmacophore DELs by assembling pre-purified sub-libraries. It is a larger self-assembled DELs than a dual-pharmacophore DELs, and each library member can be purified and characterized. The challenge in constructing trio-pharmacophore DELs is the site-directed synthesis of multivalent molecules on suitable organic scaffolds [4].
Alfa Chemistry is exploring the construction strategy of trio-pharmacophore DELs. Please click the link above to view our services in detail.
Choose Alfa Chemistry

Professional research team

Complete DELs platform

Extensive experimental experience

Strict quality control system
References
- Ma, P.; et al. Evolution of chemistry and selection technology for DNA-encoded library. Acta Pharmaceutica Sinica B. 2023.
- Adrián, G.-M.; et al. DNA-encoded chemical libraries: a comprehensive review with succesful stories and future challenges. ACS Pharmacol Transl Sci. 2021, 4(4): 1265-1279.
- Neri, D; Lerner, R. A. DNA-encoded chemical libraries: a selection system based on endowing organic compounds with amplifiable information. Annual Review of Biochemistry. 2018, 87(1): 5.1-5.24.
- Cui, M.; et al. Trio-pharmacophore DNA-encoded chemical library for simultaneous selection of fragments and linkers. Nature Communications. 2023, 14: 1481.
- Melkko, S.; et al. Lead discovery by DNA-encoded chemical libraries. Drug Discov. Today. 2007, 12: 465–471.
