Protein-protein interactions (PPIs) represent specific physical contacts between two or more protein molecules, many occurring within the intricate biomolecular environment of a cell or organism. These interactions play crucial roles in regulating diverse biological processes, including cellular functions and signaling pathways. Notably, aberrant PPIs have been implicated in various diseases such as cancer, infectious diseases, and neurodegenerative disorders.
However, traditional approaches in medicinal chemistry face significant hurdles in effectively targeting PPIs, primarily due to the large, flat interaction surfaces involved, which differ from the binding pockets of conventional protein targets [1]. Consequently, there is a pressing need to devise more efficient methodologies for screening PPI targets. In recent years, DNA encoded libraries (DELs) technology has emerged to address this challenge and identify PPI modulators.
PPI-Based Hit Identification
DELs selection process stands as a robust method for PPI-based drug discovery, leveraging affinity-based small molecule selection. With a deep understanding of the target PPI mechanism of action (MOA) and the rational design of interaction partner groups, DELs are able to deliver selective hits with the desired MOA and potentially specified functionality. Three types of hit identification based on PPI are outlined below.
- Direct PPI blocker identification: Through screening of DELs, containing a vast diversity of molecules, PPI interface binders can be directly uncovered. Subsequent validation studies on these binders facilitate the identification of PPI blockers.
- Function-oriented hit identification: Alterations in PPI protein conformation can impact PPI, consequently influencing changes in PPI ligand binding capability. DELs can be screened under conformation-based regulation to identify hits and provide extensive information on the candidate's potential function.
- Identification of specific binders with specific MOA: In some cases, the PPI network of the target may exhibit complex interactions with different partner proteins. By including all possible interaction scenarios in DELs selection and exploring the target PPI mechanism of action (MOA), the chances of identifying robust hits with specific functions are maximized.
Examples of DELs Screen for PPI
DELs emerged as a pivotal tool in identifying PPI hits, contributing to the discovery of small molecule PPI modulators capable of influencing specific classes of PPIs. Below are examples of PPI modulators or inhibitors identified through DELs selection for various PPI targets [2].
Lymphocyte Function-Associated Antigen 1 (LFA-1)
LFA-1, an integrin expressed on lymphocytes and other leukocytes, stands as an established therapeutic target for inflammatory and autoimmune diseases. Submicromolar allosteric inhibitors of LFA1 were identified from a 4.1 billion member DELs containing a central triazine core, with the most potent hit showing an IC50 of 16 nM (Fig. 1) [2].
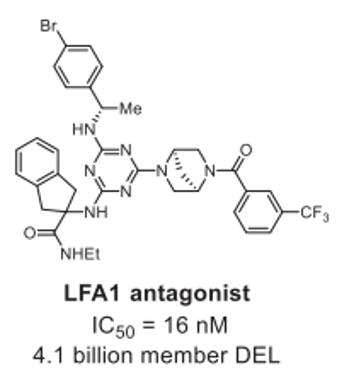 Fig. 1 LFA1 inhibitors identified from DELs [2].
Fig. 1 LFA1 inhibitors identified from DELs [2].
Myeloid Cell Leukemia 1 (Mcl-1)
Mcl-1, an anti-apoptotic BCL2 family member, presents a high-interest PPI target for hematological cancer therapy. A single-digit nanomolar inhibitor (Fig. 2) of Mcl1 was identified through DELs selection. Upon co-crystallization with Mcl1, this inhibitor demonstrated binding in a β-turn conformation, bringing its two ends into close proximity [2].
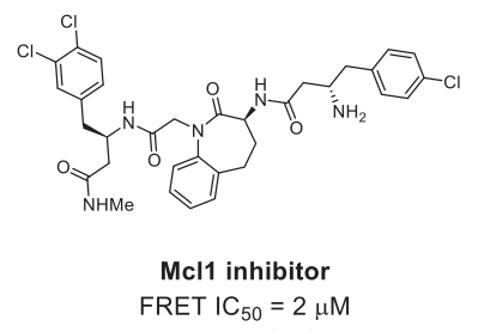 Fig. 2 Mcl1 inhibitors identified from DELs [2].
Fig. 2 Mcl1 inhibitors identified from DELs [2].
X-Linked Inhibitor of Apoptosis Protein (XIAP)
XIAP, a PPI target crucial for halting apoptosis, can also promote mitochondrial membrane permeabilization, which may provide therapeutic benefits in cancer treatments. A macrocyclic antagonist (Fig. 3) with nanomolar binding affinity was identified through DELs selection, targeting both the BIR2 and BIR3 domains of XIAP [2].
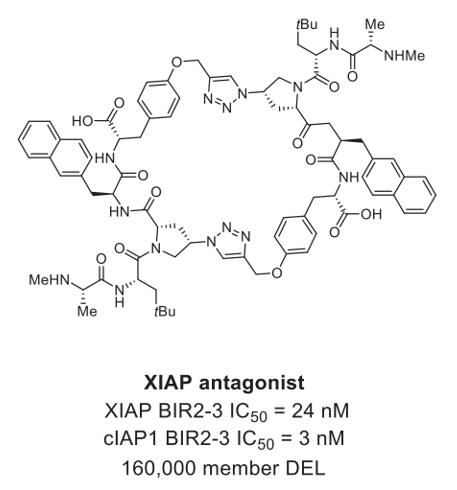 Fig. 3 XIAP antagonist identified from DELs [2].
Fig. 3 XIAP antagonist identified from DELs [2].
Advantages of DELs for Protein-Protein Interactions Targeting

- Chemical Space Exploration: DELs afford access to an unprecedented level of chemical diversity, enabling comprehensive exploration of chemical space in the context of PPI modulation.
- Rapid Hit Identification: Traditional screening approaches often suffer from limitations in throughput and scalability. DELs-based screening circumvents these challenges by leveraging the power of DNA barcoding to enable ultra-high-throughput screening of compound libraries.
- Facilitated Hit-to-Lead Optimization: Once hits are identified, DELs streamline the hit-to-lead optimization process by providing direct linkage between small molecule structures and their associated DNA barcodes.
DNA-encoded libraries (DELs) represent a paradigm shift in drug discovery, offering unprecedented opportunities for probing protein-protein interactions and identifying novel therapeutic agents. Contact us today to learn more about our services and discover how we can empower your journey towards innovation and success.
References
- Lu, H.; et al. Recent advances in the development of protein–protein interactions modulators: mechanisms and clinical trials. Signal Transduction and Targeted Therapy. 2020, 5: 213.
- Madsen, D.; et al. An overview of DNA-encoded libraries: A versatile tool for drug discovery. Prog Med Chem. 2020, 59: 181-249.
