Kinases are a diverse family of enzymes that catalyze the transfer of phosphate groups from ATP to substrates (mainly proteins or lipids). They are involved in countless cellular processes, making them indispensable for cell communication, growth and differentiation. Notably, kinases play a central role in the pathogenesis of various diseases, including cancer, vascular disease, central nervous system (CNS) disease, inflammation, and chronic inflammation.
DNA-encoded libraries (DELs) can be used to identify kinase inhibitors. In many cases, the DELs screens are able to provide non-optimized hits directly out of the library with single digit nanomolar activity on both biochemical and biophysical assays, and selectivity across other kinases [1].
Screening Strategies for Kinases Using DELs
Here are some key aspects and strategies for screening kinases using DELs:
Kinase Panel Selection
Prior to screening, it's crucial to select an appropriate panel of kinase targets relevant to the disease or pathway of interest. This panel should represent a diverse set of kinase families and isoforms.
Library Screening
DELs screening involves incubating the library with the kinase panel under conditions that promote binding interactions. After washing away unbound molecules, the DNA barcodes of the bound compounds are amplified via PCR and sequenced to identify hits.
Affinity Selection
To enrich for compounds with high affinity for the kinase targets, multiple rounds of selection can be performed. After each round, the stringency of the selection conditions can be increased to favor tighter binding interactions.
Hit Identification and Validation
Hits identified from DELs screening are typically validated using biochemical assays to confirm their binding affinity and selectivity for the target kinase. Structural biology techniques such as X-ray crystallography or cryo-electron microscopy can provide insights into the binding mode of the hit compounds.
Hit Optimization
Once validated, hit compounds can undergo medicinal chemistry optimization to improve their potency, selectivity, and pharmacokinetic properties. Structure-activity relationship (SAR) studies guided by structural biology data can inform iterative compound optimization cycles.
Functional Assays
In addition to biochemical assays, it's essential to evaluate the functional effects of the hit compounds on kinase activity using cellular or in vivo models. This helps assess their potential therapeutic relevance and mechanism of action.
Examples of DELs Screen for Kinases
Many kinase inhibitors have been discovered through DELs selection. Some of the kinases that DELs screens against and the identified kinase inhibitors are listed below.
Mitogen-activated protein (MAP) kinase
MAP kinase is one of the important signaling systems in eukaryotic cells that mediates extracellular signals to intracellular responses. P38α MAPK is an important member of the MAP kinase family and plays an important role in inflammation, various physiological and pathological processes such as cell stress, apoptosis, cell cycle and growth.
Through DELs screening of a 12.6 million member library, researchers uncovered compound VPC00628, a potent kinase inhibitor exhibiting an impressive IC50 of 7 nM in cellular assays [2] (Fig. 1).
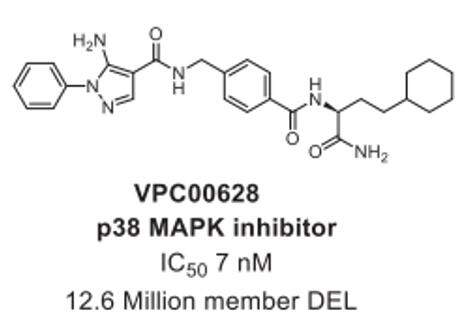 Fig. 1 p38α MAP kinase inhibitors identified from DELs [1].
Fig. 1 p38α MAP kinase inhibitors identified from DELs [1].
Aurora A Kinase
Aurora kinase is a member of the serine/threonine kinase family and is a key regulator of mitosis. Aberrant expression of Aurora kinases causes abnormal mitotic progression, leading to genetic instability and carcinogenesis, so Aurora kinase inhibitors may exert anticancer effects in various fields.
DELs screening of a 7 million member triazine library led to the discovery of Aurora A kinase inhibitors, with the most potent compound displaying an IC50 of 0.27 μM, as shown in Fig. 2 [2].
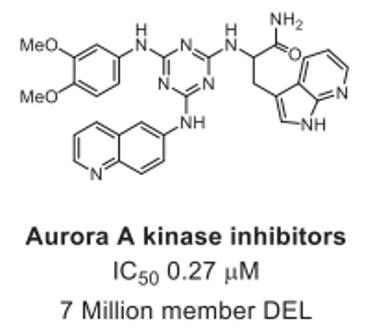 Fig. 2 Aurora A kinase inhibitors identified from DELs [1].
Fig. 2 Aurora A kinase inhibitors identified from DELs [1].
Receptor-interacting protein (RIP) kinase
RIP kinases are a family of seven members that contain a common serine-threonine kinase domain. RIP1 is a member of the RIP family that functions in a variety of cellular pathways related to both cell survival and death. It has emerged as a promising therapeutic target for inflammation, autoimmune, and neurodegenerative diseases.
DELs selection facilitated the identification of the GSK'481 hit series (2), characterized by high RIP1 potency and kinase selectivity. Subsequent optimization efforts yielded two clinical candidates, GSK3145095 (1) and GSK2982772 (3), boasting good oral bioavailability and high potency (Fig. 3) [3, 4].
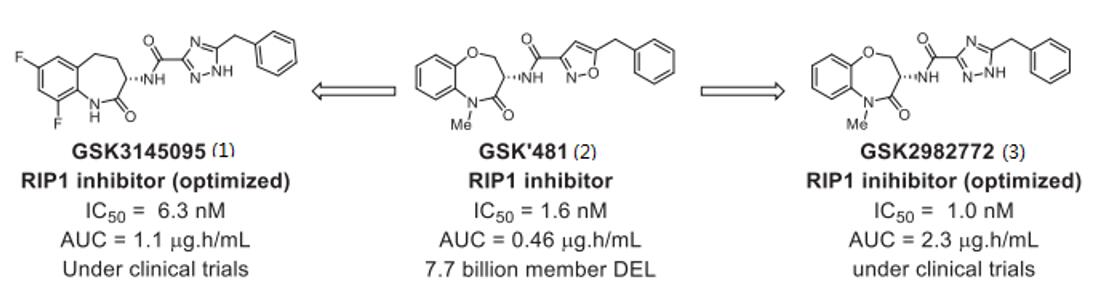 Fig. 3 RIP inhibitors identified from DELs [1].
Fig. 3 RIP inhibitors identified from DELs [1].
Advantages of DELs in Kinases Targeting

- High Throughput Screening: DELs facilitate the screening of an extensive chemical space, allowing for the rapid identification of potential kinase inhibitors.
- Efficient Hit Identification: DELs screens have yielded non-optimized hits with single-digit nanomolar activity and selectivity across multiple kinases, accelerating the lead discovery process.
- Diverse Chemical Space: With DELs comprising diverse chemical scaffolds, they offer a rich source of novel kinase inhibitors with varied mechanisms of action.
- Cost-Effective and Time-Efficient: DELs technology streamlines the hit identification process, reducing costs and accelerating the pace of drug discovery campaigns.
DELs technology has revolutionized the landscape of kinase discovery, offering unprecedented insights into kinase inhibition and paving the way for innovative therapeutic interventions. Contact us today to learn more about our services and discover how we can empower your journey towards innovation and success.
References
- Madsen, D.; et al. An overview of DNA-encoded libraries: A versatile tool for drug discovery. Prog Med Chem. 2020, 59: 181-249.
- Petersen, L. K.; et al. Novel p38α MAP kinase inhibitors identified from yoctoReactor DNA-encoded small molecule library. Med Chem Comm. 2016, 7: 1332–9.
- Harris, P. A.; et al. DNA-encoded library screening identifies benzo[b][1,4]oxazepin-4-ones as highly potent and monoselective receptor interacting protein 1 kinase inhibitors. J Med Chem. 2016, 59: 2163–78.
- Harris, P. A.; et al. Discovery of a first-in-class receptor interacting protein 1 (RIP1) kinase specific clinical candidate (GSK2982772) for the treatment of inflammatory diseases. J Med Chem. 2017, 60: 1247–61.
