After applying DNA encoded libraries (DELs) to screen small-molecule ligands for target proteins, it is necessary to perform polymerase chain reaction (PCR) amplification of the DNA barcodes of preferential protein binders, followed by high-throughput DNA sequencing (also named next-generation sequencing, NGS). For DNA sequencing results, data analysis or decoding is required to be carried out, aiming to convert the DNA sequence into the corresponding compounds with these DNA tags.
Alfa Chemistry has focused on DELs technology for many years and has full experience in this field. After DELs selection, we are able to provide data analysis services, mainly for sequencing data from PCR amplifications and NGSs. Please do not hesitate to contact us with any concerns, and we would like to serve you.
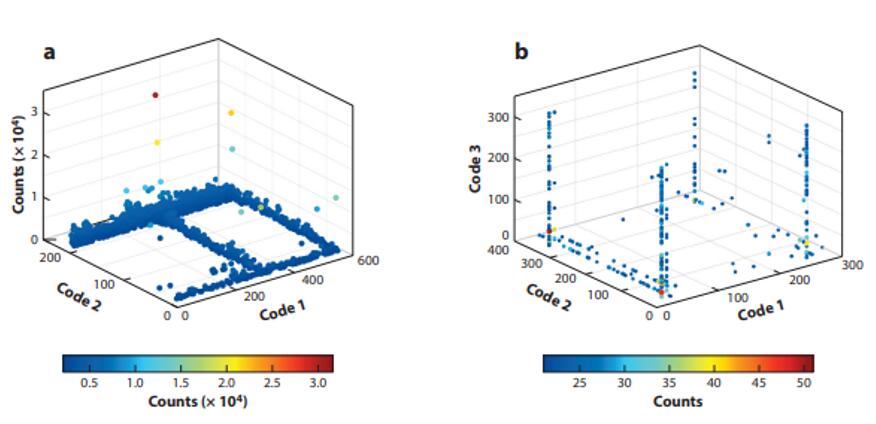 Fig. 1 Illustrative fingerprints of selection results, performed with a library consisting of (a) two or (b) three sets of building blocks [1].
Fig. 1 Illustrative fingerprints of selection results, performed with a library consisting of (a) two or (b) three sets of building blocks [1].
Challenges
When it comes to data analysis of DELs selection, false negatives and noisy data are the major challenges, especially for large DELs where library quality is compromised by truncating and/or side products during library synthesis [2]. Statistical methods developed by Alfa Chemistry addressed these issues effectively.
Principles
Decoding refers to the process of analyzing a lot of scattered data from DNA sequencing to get a conclusion of biological significance. For the DELs technique, the chemical structure of enrichment compounds can be decoded by comparing the correspondence between DNA coding and chemical building blocks (BBs) in the design of the library.
Decoding involves determining the degree of enrichment of all library members, primarily by determining the counts of sequences encoded by each compound. Considering that some false positives may be introduced during selection and sequencing, it is important to select appropriate data analysis tools and methods. The statistical evaluation of library selection output data can, in general, be represented by a suitable graphical display, such as a two- or three-dimensional plot (called "fingerprints") showing the relative abundance of each library member is. Before and after DELs selection, there are obvious differences in sequence counts between library members. Structure-activity relationships (SAR) and Lipinski's Rule of Five (Lipinski) are combined to find compounds with binding activity and improved properties. After decoding, preferential binders need to be resynthesized off-DNA and tested for affinity constants later [3].
Our Capabilities

Alfa Chemistry has a DELs technology data analysis automation platform that enables automatic data processing and reporting.
Alfa Chemistry has powerful hit identification algorithms that enables the identification of enrichment signatures from sequencing data.
Alfa Chemistry has fast analysis, with automated reporting of DELs selection results completed in approximately one day.
Alfa Chemistry has rich experience, allowing professional bioinformatics experts ready to communicate with customers on plans.
Order Process
References
- Neri, D; Lerner, R. A. DNA-encoded chemical libraries: a selection system based on endowing organic compounds with amplifiable information. Annual Review of Biochemistry. 2018, 87(1): 5.1-5.24.
- Ma, P.; et al. Evolution of chemistry and selection technology for DNA-encoded library. Acta Pharmaceutica Sinica B. 2024, 14(2): 492-516.
- Favalli, N.; et al. DNA-encoded chemical libraries – achievements and remaining challenges. FEBS Letters. 2018, 592(12): 2168-2180.
