Proteases are enzymes that cleave peptide bonds in proteins, playing critical roles in numerous physiological processes and disease states. DNA-encoded libraries (DELs) are particularly valuable in the discovery of inhibitors and modulators for proteases due to their ability to screen vast numbers of compounds efficiently.
Application of DELs in Protease Discovery
DELs in the discovery of proteases can through the following types:
High-Throughput Screening
- Large Library Size: DELs allow the screening of billions of compounds simultaneously, significantly enhancing the chances of finding potent and selective protease inhibitors.
- Efficient Screening Process: The use of DNA tags facilitates rapid identification and quantification of binding events, making the screening process both time- and cost-effective.
Target-Specific Libraries
- Focused Libraries: Libraries can be designed with specific chemical scaffolds known to interact with proteases, increasing the likelihood of identifying effective inhibitors.
- Iterative Screening: DELs allow for iterative rounds of selection and amplification, refining the pool of potential inhibitors with each round.
Identification of Novel Inhibitors
- Diverse Chemical Space: DELs explore a wide range of chemical space, potentially identifying novel chemotypes that traditional libraries might miss.
- Non-Standard Inhibitors: DELs can identify non-standard or allosteric inhibitors that bind to protease sites other than the active site, offering new therapeutic strategies.
Specific Applications in Disease-Related Proteases
Cancer
- Matrix Metalloproteinases (MMPs): DELs can be used to discover inhibitors for MMPs, which are involved in the breakdown of extracellular matrix components and are implicated in cancer metastasis.
- Caspases: Targeting caspases involved in apoptosis regulation can lead to novel cancer therapies.
Infectious Diseases
- Viral Proteases: Proteases from viruses like HIV, HCV, and SARS-CoV-2 are critical for viral replication. DELs can be used to find inhibitors that block these proteases, hindering virus maturation and proliferation.
- Bacterial Proteases: Inhibitors for bacterial proteases involved in virulence can be identified using DELs, offering new antibacterial strategies.
Inflammatory Diseases
- Cathepsins: These lysosomal proteases are involved in various inflammatory processes. DELs can help discover inhibitors to modulate these enzymes' activity, offering potential treatments for conditions like arthritis and autoimmune diseases.
Neurodegenerative Diseases
- Calpains: These calcium-dependent cysteine proteases are implicated in neurodegenerative diseases like Alzheimer's. DELs can identify inhibitors that may prevent neurodegeneration.
Examples of DELs Screen for Proteases
Below are examples of proteases identified through DELs selection:
SARS-CoV-2 Main Protease
Recent applications of DELs in response to the COVID-19 pandemic have led to the rapid identification of inhibitors targeting the main protease of SARS-CoV-2, crucial for viral replication [1].
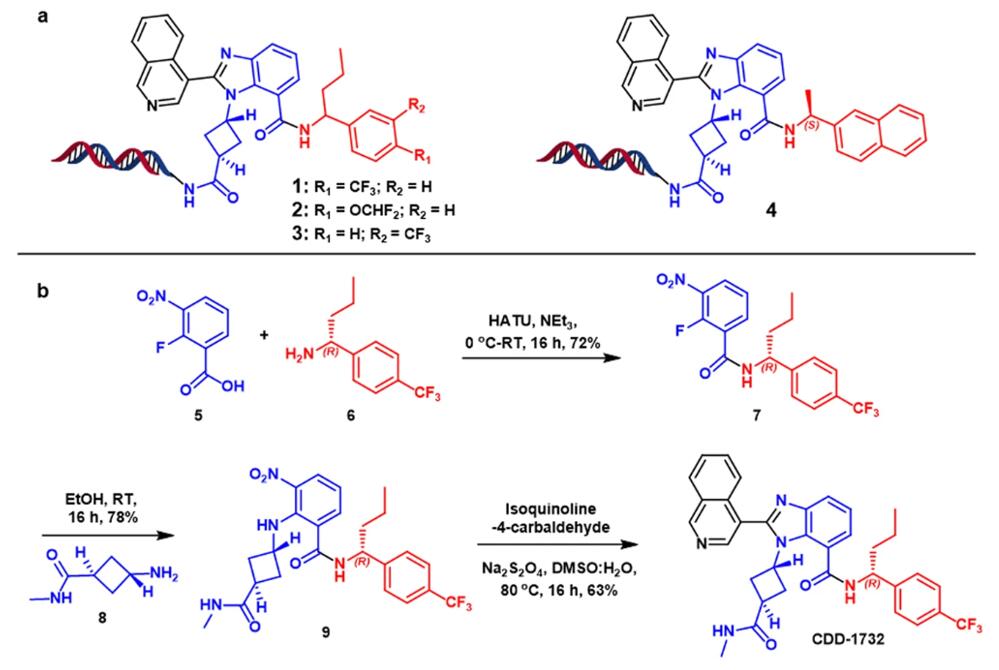 Fig. 1 DNA-encoded libraries selections of SARS-CoV-2 Mpro
Fig. 1 DNA-encoded libraries selections of SARS-CoV-2 Mpro
a. Hit compounds enriched from selections. b Off-DNA synthesis of CDD-1732.
HIV Protease Inhibitors
DELs technology has been used to identify novel inhibitors against HIV protease, showcasing the potential of DELs in antiviral drug discovery.
Advantages of Using DELs for Protease Targeting

- Scalability: The ability to screen billions of compounds provides a significant advantage over traditional methods.
- Speed: Rapid identification and optimization of hits due to the high-throughput nature of DELs screening.
- Cost-Effectiveness: Reduced cost per compound screened compared to conventional high-throughput screening methods.
- Automation: The processes involved in DELs screening are highly amenable to automation, further increasing efficiency.
DELs are a powerful tool in the identification and development of protease inhibitors, offering significant advantages in terms of diversity, efficiency, and cost-effectiveness. Contact us today to learn more about our services and discover how we can empower your journey towards innovation and success.
Reference
- Jimmidi R, Chamakuri S, Lu S, et al. DNA-encoded chemical libraries yield non-covalent and non-peptidic SARS-CoV-2 main protease inhibitors. Commun Chem. 2023 Aug 4;6(1):164.
