Enzymes play a crucial role in various drug applications due to their unique catalytic properties and specificity. DNA-encoded libraries (DELs) have emerged as a powerful technology in drug discovery, offering a unique approach to identifying potent ligands for various biological targets, including enzymes.
Application of DELs in Enzyme Targeting
DELs offer a highly efficient and scalable approach to identifying small molecule ligands for various biological targets, including enzymes. Here are the applications of DELs in enzyme targeting:
High-Throughput Screening
DELs enable the simultaneous screening of billions of compounds against enzyme targets. This high-throughput capability is particularly advantageous for enzymes, which are crucial targets in drug discovery due to their role in catalyzing biochemical reactions.
Structure-Activity Relationship (SAR) Studies
DELs facilitate the rapid generation of SAR data by allowing researchers to screen a diverse array of chemical scaffolds. The DNA tags help in tracking the interaction between specific compounds and enzymes, thereby accelerating the identification of active molecules and optimization of their properties.
Mechanism of Action Studies
By using DELs, researchers can explore the mechanism of action of enzyme inhibitors. DNA encoding allows for precise identification of binding sites and interaction patterns, which helps in understanding how these inhibitors affect enzyme activity.
Target Validation
DELs can assist in validating new enzyme targets by identifying small molecules that modulate enzyme activity. This validation is crucial in early-stage drug discovery to ensure that targeting a particular enzyme will yield therapeutic benefits.
Examples of DELs Screen for Enzymes
Below are examples of enzymes identified through DELs selection:
NAD+‑Dependent Enzymes
DELs offer a revolutionary approach to discovering small molecule ligands for various biological targets, including NAD+ (nicotinamide adenine dinucleotide) dependent enzymes. These enzymes play critical roles in cellular metabolism, signaling, and homeostasis, making them attractive targets for drug discovery. Yuen [1] introduced the first DELs designed to target NAD+ -binding pockets. This library contains a diverse array of compounds and was investigated for its utility in sampling the chemical binder space of NAD+ -dependent enzymes.
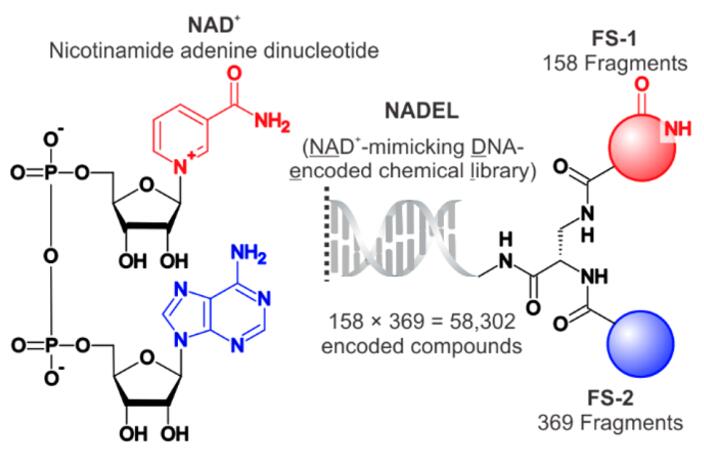 Fig. 1 Structure of NADEL and relationship to NAD+ [1].
Fig. 1 Structure of NADEL and relationship to NAD+ [1].
Sirtuin modulators
Sirtuins (SIRT), includes seven isoforms (SIRT1-7), are enzymes reliant on NAD+ that function as protein lysine deacylases and mono-ADP ribosylases. Consequently, targeting sirtuin activity has emerged as a promising therapeutic strategy for numerous pathologies. Zhou et al. [2] recently discovered compound 77-39 as a SIRT3 inhibitor (IC50 = 4.5 μM) through screening a DNA-encoded library.
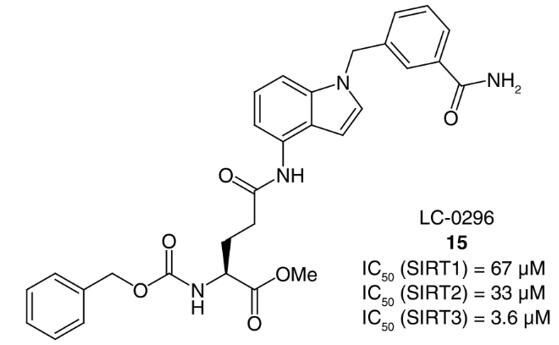 Fig. 2 Structures of SIRT3 [2].
Fig. 2 Structures of SIRT3 [2].
Advantages of DELs in Enzyme Research

- Diversity and Scale: DELs allow the creation of highly diverse libraries that cover a broad chemical space, increasing the likelihood of finding hits against enzyme targets.
- Efficiency: The DNA-encoding method enables rapid synthesis and screening, significantly reducing the time and cost compared to traditional drug discovery methods.
- Precision: DNA barcoding provides precise identification and tracking of active compounds, allowing for detailed analysis and optimization.
- Flexibility: DELs can be adapted to a wide range of enzyme targets, including those considered challenging or "undruggable" by conventional approaches.
DELs represent a transformative approach in the discovery and development of enzyme inhibitors, offering a highly efficient and versatile platform for identifying new therapeutic agents. Contact us today to learn more about our services and discover how we can empower your journey towards innovation and success.
References
- Yuen LH, Dana S, Liu Y, et al. A Focused DNA-Encoded Chemical Library for the Discovery of Inhibitors of NAD+-Dependent Enzymes. J Am Chem Soc. 2019 Apr 3;141(13):5169-5181.
- Zhou Y, Li C, Peng J et al. DNA-encoded dynamic chemical library and its applications in ligand discovery. J. Am. Chem. Soc. 2018, 140(46), 15859–15867.
